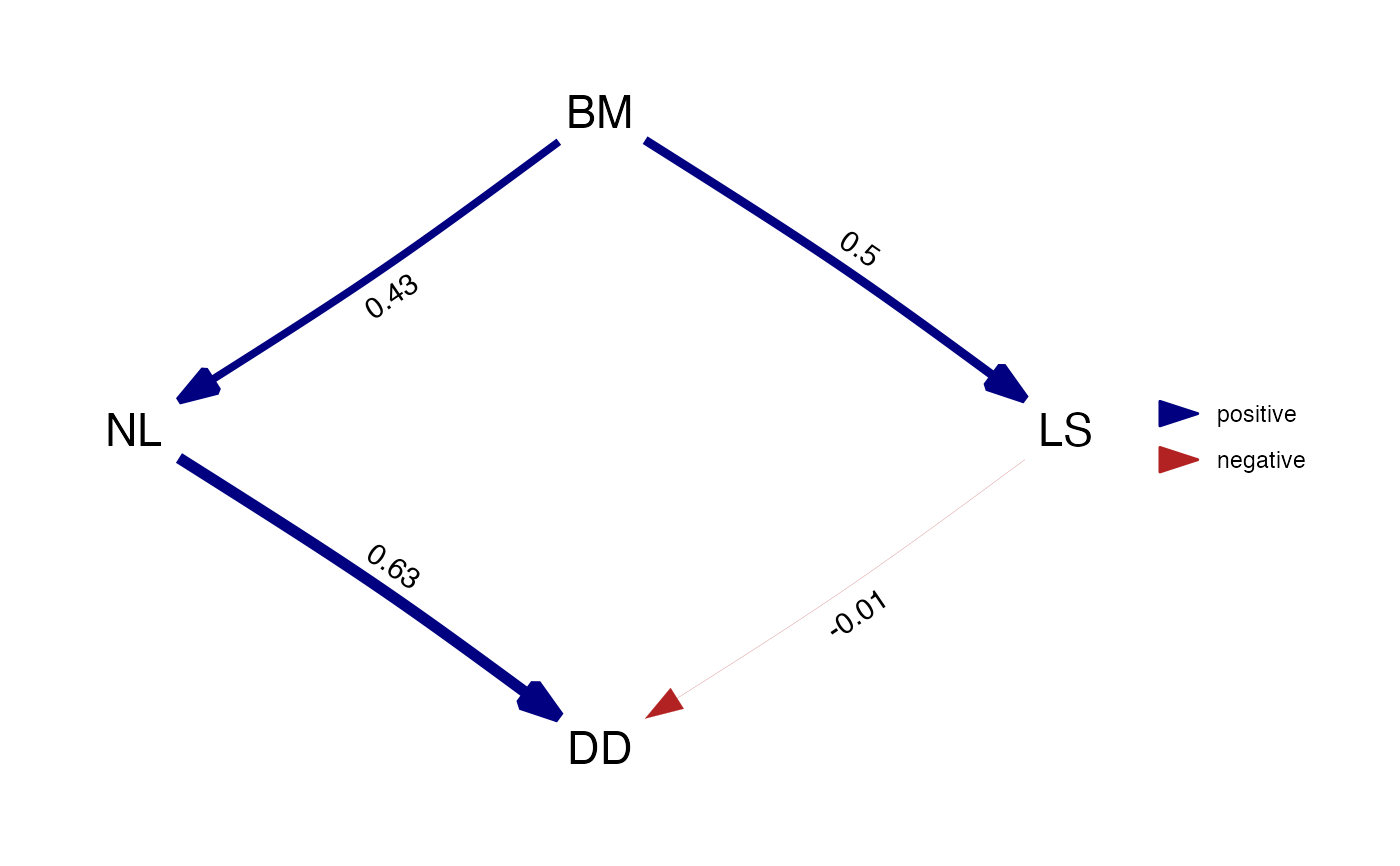
Add standardized path coefficients to a DAG.
est_DAG(DAG, data, tree, model, method, boot = 0, ...)Arguments
- DAG
A directed acyclic graph, typically created with
DAG.- data
A
data.framewith data. If you have binary variables, make sure they are either character values or factors!- tree
A phylogenetic tree of class
phylo.- model
The evolutionary model used for the regressions on continuous variables. See phylolm::phylolm for options and details. Defaults to Pagel's lambda model
- method
The estimation method for the binary models. See phylolm::phyloglm for options and details. Defaults to logistic MPLE.
- boot
The number of bootstrap replicates used to estimate confidence intervals.
- ...
Arguments passed on to
phylolm:lower.bound: optional lower bound for the optimization of the phylogenetic model parameter.upper.bound: optional upper bound for the optimization of the phylogenetic model parameter.starting.value: optional starting value for the optimization of the phylogenetic model parameter.measurement_error: a logical value indicating whether there is measurement error sigma2_error (see Details).Arguments passed on to
phyloglm:btol: bound on the linear predictor to bound the searching space.log.alpha.bound: bound for the log of the parameter alpha.start.beta: starting values for beta coefficients.start.alpha: starting values for alpha (phylogenetic correlation).
Value
An object of class fitted_DAG.